Ensuring Output Consistency of Distributed Machine Learning Algorithms

During the productionization process of our machine learning patterns, we observed variations in our model predictions and probability score outputs when executed in Spark on a distributed cluster environment. This variation in model outputs occur when model type, hyperparameters, seed value and input data are held constant. After rigorous experimentations and analysis, we determined a method which guarantees consistency in the model outputs regardless of cluster configuration, platform (EMR vs Databricks) and data format (eg. BZ, ORC, PARQUET).
At a high level, our experiments consisted of running our models on several cluster configurations which included different Spark versions, instance types, number of instances and autoscaling. Below were our initial observations:
- Model produced consistent results when maintaining the same cluster configuration for each run.
- For small input datasets (~ <= 16 mb), results are consistent regardless of cluster configuration.
- For large input datasets, results are inconsistent across multiple cluster configurations.
These preliminary observations led us into taking a deeper dive into some of the Spark internals through which we were able to narrow in and discover the root cause for our model output variations.
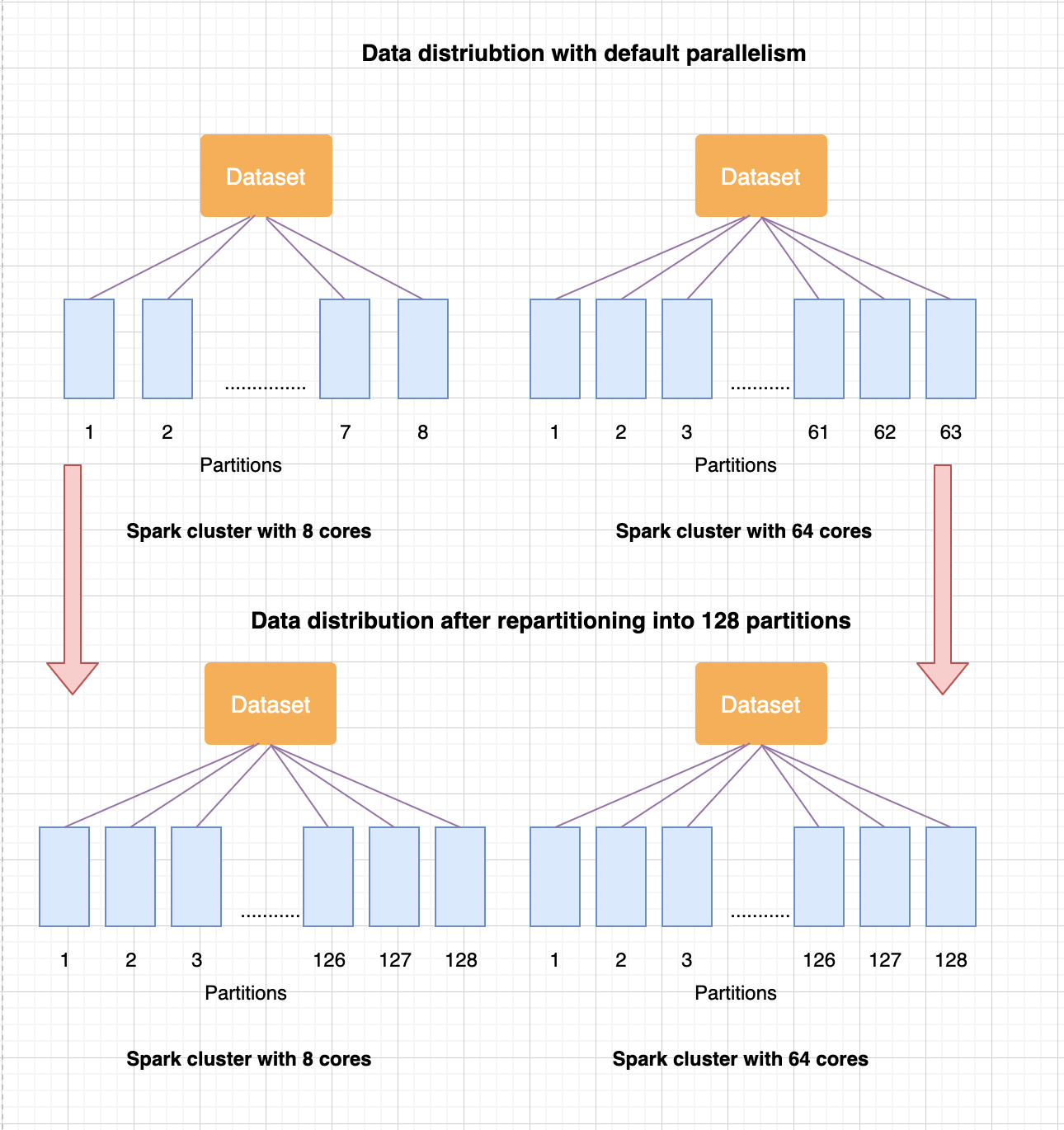
When Spark reads in an input dataset, the data internally gets split into partitions and executes computations on those partitions in parallel. Spark uses default parallelism (when not set by user) to decide the number of partitions. The number of partitions created by Spark correlates to the number of cores available across all worker nodes in a cluster. When the cluster configuration is changed, the data distribution and ordering of the partition data also gets changed and thus impacts the machine learning algorithm output.
The solution to address the issue is to ensure the input data is distributed and ordered in a consistent manner irrespective of cluster configuration. To achieve this, equally partition the dataset using a key column(s) and perform a local sort within each partition using same key column(s) to maintain the order. When repartitioning the data, the number of partitions configured should be equal to or a multiple of the number of cores (2 or 3 times) in the cluster. Perform this data preparation step prior to executing machine learning algorithm.
The following diagram illustrates partitioning, and the differences between default versus repartitioning states:
Example 1 – Using previously split and saved input dataset (Split into Train and Test sets)
//Read saved input datasets
val repartTrainSetDf = trainSetDf.repartition(numPartition, col(“key Column”)).sortWithinPartitions(“key Column”)
// Note: as the test dataset is only required for evaluating the model, it is not required to repartition the test dataset.
// Train the model
// Evaluate the model with test dataset
Example 2 – Using randomSplit to split the input dataset into training and test sets
//Read input dataset
repartTrainingDataDf=trainingDataDf.repartition(numPartition,col(“key column”)).sortWithinPartitions(“key Column”)
val (trainSetDf, testSetDf) = repartTrainingDataDf.randomSplit(Array(0.80, 0.20), seed = 1)
// Train the model
// Evaluate the model with test dataset
In the above examples, the key column(s) are the column(s) which uniquely identify each record in the dataset. In the case the dataset does not already contain a unique key column(s), then one approach you can use is the Spark provided function “monotonically_increasing_id()” which is optimized for performance and generates a unique id for each row. Alternatively, you can use the window function “row_number” to generate unique row number (row_number() over (order by col1, col2…). As stated earlier, the number of partitions (numPartitions), should be equal to or multiples of number of cores available in the cluster.
Note that the provided solution may not work if autoscaling is turned ON in the cluster. This is because the autoscaling will re-shuffle the data and the predefined order set by repartition and sort may get disturbed and hence the training algorithm may yield different outputs.
Also, it is recommended to keep track of the cluster configuration, hyper parameters and seed values used for the model training.
